Research Overview
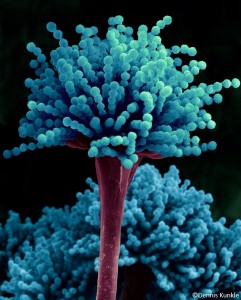 The broad goal in our laboratory is to better understand, and then beneficially manipulate, microbial expression systems. While in the past we have carried out projects involving bacteria, all our current work is with filamentous fungi. Specifically, we use fungi to better understand eukaryotic gene-regulatory networks. Our primary focus is on filamentous fungal morphogenesis and cell wall biosynthesis. We use a multi-omics approach to understand systems behaviors. Phosphoproteomic analysis informs us regarding kinase-mediated signal transduction, and transcriptomics/proteomics informs us regarding cellular outcomes. These approaches allow us to develop hypotheses regarding cell function, which are tested using both a genetic approach and a sophisticated set of analytical tools (electron microscopy, digital image analysis, atomic force microscopy) to asses fungal morphology and the physical properties of fungal cell walls.
The broad goal in our laboratory is to better understand, and then beneficially manipulate, microbial expression systems. While in the past we have carried out projects involving bacteria, all our current work is with filamentous fungi. Specifically, we use fungi to better understand eukaryotic gene-regulatory networks. Our primary focus is on filamentous fungal morphogenesis and cell wall biosynthesis. We use a multi-omics approach to understand systems behaviors. Phosphoproteomic analysis informs us regarding kinase-mediated signal transduction, and transcriptomics/proteomics informs us regarding cellular outcomes. These approaches allow us to develop hypotheses regarding cell function, which are tested using both a genetic approach and a sophisticated set of analytical tools (electron microscopy, digital image analysis, atomic force microscopy) to asses fungal morphology and the physical properties of fungal cell walls.
Systems Biology
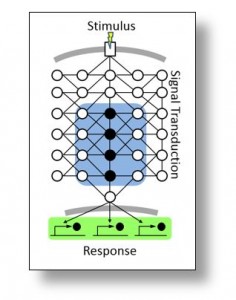
The goal of this project is to develop a new approach for modeling gene regulatory networks. We are testing the hypothesis that initial experimental characterization of a network subset will permit identification of the biomolecular constituents and their connectivity, thus establishing network topology. System wide time-course measurements can then be used to refine this network into a reaction kinetic model capable of making accurate system predictions. The cell wall integrity signaling pathway of the experimentally tractable model fungus Aspergillus nidulans is serving as a model. This pathway responds to cell wall damage by activating repair mechanisms that restore cell integrity. Because protein kinases play a pivotal role in mediating cellular regulatory activities, we are focusing on a subset of kinases and the discovery of their associated substrates to initially assemble a rudimentary network. In addition, the system is being experimentally perturbed, so we can measure its dynamic response using a robust proteomic and phosphoproteomic platform. This project involves collaboration with both Iowa State University and the University of Connecticut and is sponsored by the National Science Foundation.
Mycelial Materials
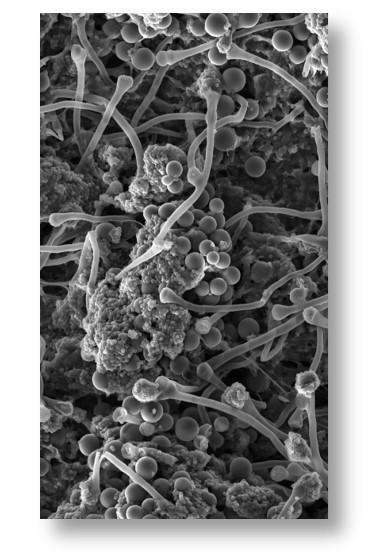 The vast majority of the world’s manufactured materials are nonrenewable and energy-intensive to produce. Viable alternatives must be developed, and “mycelial materials” made from filamentous fungi are an exciting option. Compared to traditional materials, these have lower raw-material cost, require less energy to produce, and are eminently biodegradable. And mycelial materials can have a diverse range of mechanical properties (e.g., similar to leather, foam, wood) due to the complex morphology of filamentous fungi (image to the left). However, the current approach to modify mycelial-material mechanical properties is archaic, and is typically done by trial-and-error using different fungal species, altering nutrients used for growth, or changing the growth environment. Because it’s unclear how these inputs lead to specific changes in material properties, advances have been limited. We are addressing these limitations by developing an in-depth understanding of the relationships between genetic variables, fungal morphological phenotypes, and mycelial-material mechanical properties. To do this we are leveraging findings from our systems biology work (above) related to morphogenesis and cell-wall biosynthesis. This work is being carried out by our collaborative team, who have expertise in diverse disciplines (e.g., fungal genetics, fungal cell biology, materials science/analysis, and machine learning).
The vast majority of the world’s manufactured materials are nonrenewable and energy-intensive to produce. Viable alternatives must be developed, and “mycelial materials” made from filamentous fungi are an exciting option. Compared to traditional materials, these have lower raw-material cost, require less energy to produce, and are eminently biodegradable. And mycelial materials can have a diverse range of mechanical properties (e.g., similar to leather, foam, wood) due to the complex morphology of filamentous fungi (image to the left). However, the current approach to modify mycelial-material mechanical properties is archaic, and is typically done by trial-and-error using different fungal species, altering nutrients used for growth, or changing the growth environment. Because it’s unclear how these inputs lead to specific changes in material properties, advances have been limited. We are addressing these limitations by developing an in-depth understanding of the relationships between genetic variables, fungal morphological phenotypes, and mycelial-material mechanical properties. To do this we are leveraging findings from our systems biology work (above) related to morphogenesis and cell-wall biosynthesis. This work is being carried out by our collaborative team, who have expertise in diverse disciplines (e.g., fungal genetics, fungal cell biology, materials science/analysis, and machine learning).
One of Dr. Marten’s favorite articles…
The Student, The Fish, and Agassiz
